Difference between revisions of "Cls"
| Line 25: | Line 25: | ||
<v:imagedata src="file:///C:\DOCUME~1\hkuehn\LOCALS~1\Temp\msohtml1\01\clip_image001.gif" | <v:imagedata src="file:///C:\DOCUME~1\hkuehn\LOCALS~1\Temp\msohtml1\01\clip_image001.gif" | ||
o:href="images/window-phenotypes.gif" /> | o:href="images/window-phenotypes.gif" /> | ||
| − | </v:shape><![endif]--><!--[if !vml]--> | + | </v:shape><![endif]--><!--[if !vml]-->[[image: window-phenotypes.gif]]<!--[endif]--></p> |
<p class="MsoListBullet"><!--[if !supportLists]--><span style="">●<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> </span></span><!--[endif]-->Select source file. Select a <a href="#_Phenotype_Labels_1">phenotype labels file</a> from the drop-down list. If the file is not listed, you have not yet loaded it; see <a href="#_Loading_Data_1">Loading Data</a>. GSEA updates the window to display the phenotypes in the file. Select one or more phenotypes to analyze and click <em>OK</em>. To select multiple phenotypes, use SHIFT-click or CTRL-click.</p> | <p class="MsoListBullet"><!--[if !supportLists]--><span style="">●<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> </span></span><!--[endif]-->Select source file. Select a <a href="#_Phenotype_Labels_1">phenotype labels file</a> from the drop-down list. If the file is not listed, you have not yet loaded it; see <a href="#_Loading_Data_1">Loading Data</a>. GSEA updates the window to display the phenotypes in the file. Select one or more phenotypes to analyze and click <em>OK</em>. To select multiple phenotypes, use SHIFT-click or CTRL-click.</p> | ||
<p class="MsoListBullet"><!--[if !supportLists]--><span style="">●<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> </span></span><!--[endif]-->Show phenotypes from all source files. GSEA updates the window to display all phenotypes in all files in the <em>Select source file</em> drop-down list. This is useful when you want to select phenotypes from multiple phenotype labels files. GSEA updates the window to display the phenotypes in the file. Select one or more phenotypes to analyze and click <em>OK</em>. To select multiple phenotypes, use SHIFT-click or CTRL-click.</p> | <p class="MsoListBullet"><!--[if !supportLists]--><span style="">●<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> </span></span><!--[endif]-->Show phenotypes from all source files. GSEA updates the window to display all phenotypes in all files in the <em>Select source file</em> drop-down list. This is useful when you want to select phenotypes from multiple phenotype labels files. GSEA updates the window to display the phenotypes in the file. Select one or more phenotypes to analyze and click <em>OK</em>. To select multiple phenotypes, use SHIFT-click or CTRL-click.</p> | ||
| Line 35: | Line 35: | ||
<v:imagedata src="file:///C:\DOCUME~1\hkuehn\LOCALS~1\Temp\msohtml1\01\clip_image002.gif" | <v:imagedata src="file:///C:\DOCUME~1\hkuehn\LOCALS~1\Temp\msohtml1\01\clip_image002.gif" | ||
o:href="images/window-phenotypes_onthefly.gif" /> | o:href="images/window-phenotypes_onthefly.gif" /> | ||
| − | </v:shape><![endif]--><!--[if !vml]--> | + | </v:shape><![endif]--><!--[if !vml]-->[[image: window-phenotypes_onthefly.gif]]<!--[endif]--></p> |
<p class="MsoListBullet"><!--[if !supportLists]--><span style="">●<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> </span></span><!--[endif]-->Use a gene as the phenotype. Displays the following window, which allows you to create a continuous phenotype label. Select your dataset. GSEA updates the window to display the genes in the dataset. Select a gene and click <em>Apply to dataset</em>. GSEA confirms that the gene is in the selected dataset and creates a phenotype labels file (<var>genename</var><code><span style="font-size: 10pt;">_in_</span></code><var>datasetname</var><code><span style="font-size: 10pt;">.cls</span></code>) in the default output folder. When you close this window, the new phenotype labels file appears in the <em>Select source file</em> drop-down list of the Select One or More Phenotypes window.</p> | <p class="MsoListBullet"><!--[if !supportLists]--><span style="">●<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> </span></span><!--[endif]-->Use a gene as the phenotype. Displays the following window, which allows you to create a continuous phenotype label. Select your dataset. GSEA updates the window to display the genes in the dataset. Select a gene and click <em>Apply to dataset</em>. GSEA confirms that the gene is in the selected dataset and creates a phenotype labels file (<var>genename</var><code><span style="font-size: 10pt;">_in_</span></code><var>datasetname</var><code><span style="font-size: 10pt;">.cls</span></code>) in the default output folder. When you close this window, the new phenotype labels file appears in the <em>Select source file</em> drop-down list of the Select One or More Phenotypes window.</p> | ||
<p class="MsoListContinue"><!--[if gte vml 1]><v:shape id="_x0000_i1027" type="#_x0000_t75" | <p class="MsoListContinue"><!--[if gte vml 1]><v:shape id="_x0000_i1027" type="#_x0000_t75" | ||
| Line 41: | Line 41: | ||
<v:imagedata src="file:///C:\DOCUME~1\hkuehn\LOCALS~1\Temp\msohtml1\01\clip_image003.gif" | <v:imagedata src="file:///C:\DOCUME~1\hkuehn\LOCALS~1\Temp\msohtml1\01\clip_image003.gif" | ||
o:href="images/window-phenotypes-gene.gif" /> | o:href="images/window-phenotypes-gene.gif" /> | ||
| − | </v:shape><![endif]--><!--[if !vml]--> | + | </v:shape><![endif]--><!--[if !vml]-->[[image: window-phenotypes-gene.gif]]<!--[endif]--></p> |
Revision as of 17:37, 21 March 2006
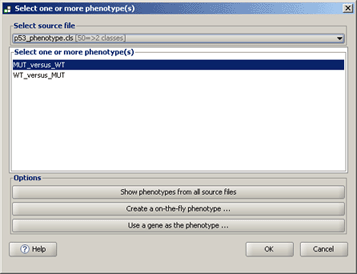
On the Run GSEA, next to the Phenotype labels parameter, click the ellipse (…) button to display the following window, which allows you to select one or more <a href="#_Phenotype_Labels_1">phenotypes</a> to analyze.
● Select source file. Select a <a href="#_Phenotype_Labels_1">phenotype labels file</a> from the drop-down list. If the file is not listed, you have not yet loaded it; see <a href="#_Loading_Data_1">Loading Data</a>. GSEA updates the window to display the phenotypes in the file. Select one or more phenotypes to analyze and click OK. To select multiple phenotypes, use SHIFT-click or CTRL-click.
● Show phenotypes from all source files. GSEA updates the window to display all phenotypes in all files in the Select source file drop-down list. This is useful when you want to select phenotypes from multiple phenotype labels files. GSEA updates the window to display the phenotypes in the file. Select one or more phenotypes to analyze and click OK. To select multiple phenotypes, use SHIFT-click or CTRL-click.
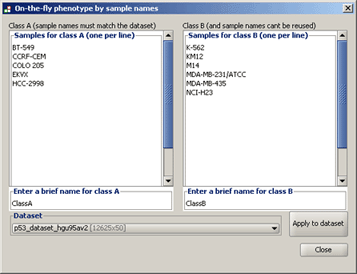
● Create an on-the-fly phenotype. Displays the following window, which allows you to create categorical phenotype labels. Enter the names of <a name="OLE_LINK4"></a><a name="OLE_LINK3">one or more samples in the box on the left and enter a name for that phenotype class (by default, ClassA). </a>Enter the names of one or more samples in the box on the right and enter a name for that phenotype class (by default, ClassB). If your dataset contains samples that you did not include in the two phenotypes, GSEA automatically excludes them from the gene set enrichment analysis of these phenotypes.
Select your dataset and click Apply to dataset. GSEA confirms that all of the samples that you specified are in the selected dataset and creates a phenotype labels file (ClassAvsClassB.cls) in the default output folder. When you close this window, the new phenotype labels file appears in the Select source file drop-down list of the Select One or More Phenotypes window.
Tip: For easier entering of sample names, cut and paste from your dataset file.
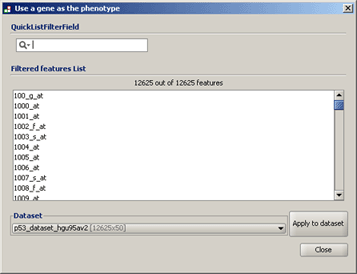
● Use a gene as the phenotype. Displays the following window, which allows you to create a continuous phenotype label. Select your dataset. GSEA updates the window to display the genes in the dataset. Select a gene and click Apply to dataset. GSEA confirms that the gene is in the selected dataset and creates a phenotype labels file (genename_in_datasetname.cls) in the default output folder. When you close this window, the new phenotype labels file appears in the Select source file drop-down list of the Select One or More Phenotypes window.