Difference between revisions of "GSEA v2.08. Release Notes"
m |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | [http://www.broadinstitute.org/gsea/ GSEA Home] | | ||
| + | [http://www.broadinstitute.org/gsea/downloads.jsp Downloads] | | ||
| + | [http://www.broadinstitute.org/gsea/msigdb/ Molecular Signatures Database] | | ||
| + | [http://www.broadinstitute.org/cancer/software/gsea/wiki/index.php/Main_Page Documentation] | | ||
| + | [http://www.broadinstitute.org/gsea/contact.jsp Contact] <br /> | ||
| + | <br /> | ||
<p>In release 2.0.8, there is <strong>no change</strong> to the GSEA algorithm.</p> | <p>In release 2.0.8, there is <strong>no change</strong> to the GSEA algorithm.</p> | ||
<p>The changes include:</p> | <p>The changes include:</p> | ||
<ol> | <ol> | ||
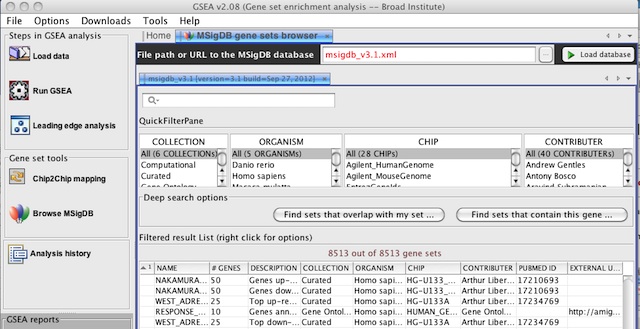
| − | <li> | + | <li>When you Browse MSigDB, by default it will now display version 3.1 of the MSigDB gene sets. |
| − | <p>To browse MSigDB | + | <p>To browse earlier versions of MSigDB, type <strong>msigdb_v3.xml</strong> or <strong>msigdb_v2.5.xml</strong> in the <em>File path or URL to the MSigDB database</em> and click the <em>Load database</em> button.</p> |
<p>[[Image:Loading MSigDB3.1.jpg]]</p> | <p>[[Image:Loading MSigDB3.1.jpg]]</p> | ||
| + | <br> | ||
</li> | </li> | ||
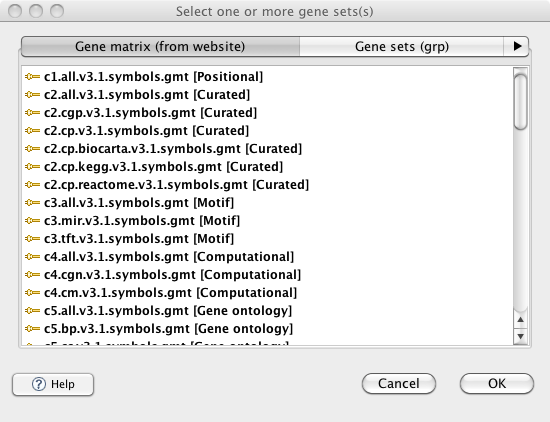
| − | <li> | + | <li><p>In the previous version of GSEA, there were display issues when viewing the selection menu of GMT files in the MSigDB database from the <em>Run Gsea</em> screen and the <em>Chip2Chip mapping</em> screen. These issues have been fixed. The GMT files for the latest release of MSigDB are now always displayed at the top of the list and in a bold font.</p> |
| + | <p>[[Image:Viewing_GMT_files.png]]</p> | ||
</li> | </li> | ||
| − | <li> | + | <li><p>The latest version of MSigDB is significantly larger than previous versions and we now recommend that you run GSEA with no less than 1 GB of memory. If you run GSEA from the command line, use the -Xmx Java option to specify increased heap size. For example, <tt>java -Xmx1024m -jar gsea2-2.08.jar</tt>.</p> |
| − | <p>The | ||
</li> | </li> | ||
</ol> | </ol> | ||
Latest revision as of 03:16, 25 September 2016
GSEA Home |
Downloads |
Molecular Signatures Database |
Documentation |
Contact
In release 2.0.8, there is no change to the GSEA algorithm.
The changes include:
- When you Browse MSigDB, by default it will now display version 3.1 of the MSigDB gene sets.
To browse earlier versions of MSigDB, type msigdb_v3.xml or msigdb_v2.5.xml in the File path or URL to the MSigDB database and click the Load database button.
In the previous version of GSEA, there were display issues when viewing the selection menu of GMT files in the MSigDB database from the Run Gsea screen and the Chip2Chip mapping screen. These issues have been fixed. The GMT files for the latest release of MSigDB are now always displayed at the top of the list and in a bold font.
The latest version of MSigDB is significantly larger than previous versions and we now recommend that you run GSEA with no less than 1 GB of memory. If you run GSEA from the command line, use the -Xmx Java option to specify increased heap size. For example, java -Xmx1024m -jar gsea2-2.08.jar.