Difference between revisions of "GSEA v2.0.7 Release Notes"
(added toggling) |
m |
||
| Line 1: | Line 1: | ||
| + | <a href="http://www.broadinstitute.org/gsea/">GSEA Home</a> | <a href="http://www.broadinstitute.org/gsea/downloads.jsp">Downloads</a> | <a href="http://www.broadinstitute.org/gsea/msigdb/">Molecular Signatures Database</a> | <a href="http://www.broadinstitute.org/cancer/software/gsea/wiki/index.php/Main_Page">Documentation</a> | <a href="http://www.broadinstitute.org/gsea/contact.jsp">Contact</a> <br /> | ||
| + | <br /> | ||
In release 2.0.7, there is <strong>no change</strong> to the GSEA algorithm.<br /> | In release 2.0.7, there is <strong>no change</strong> to the GSEA algorithm.<br /> | ||
<br /> | <br /> | ||
Revision as of 10:13, 11 April 2013
<a href="http://www.broadinstitute.org/gsea/">GSEA Home</a> | <a href="http://www.broadinstitute.org/gsea/downloads.jsp">Downloads</a> | <a href="http://www.broadinstitute.org/gsea/msigdb/">Molecular Signatures Database</a> | <a href="http://www.broadinstitute.org/cancer/software/gsea/wiki/index.php/Main_Page">Documentation</a> | <a href="http://www.broadinstitute.org/gsea/contact.jsp">Contact</a>
In release 2.0.7, there is no change to the GSEA algorithm.
However, in the MSigDB browser, you can now choose which version of MSigDB you want to load into GSEA. By default, the MSigDB browser points to v3.0 of MSigDB, and you can load this information by clicking the Load database button.
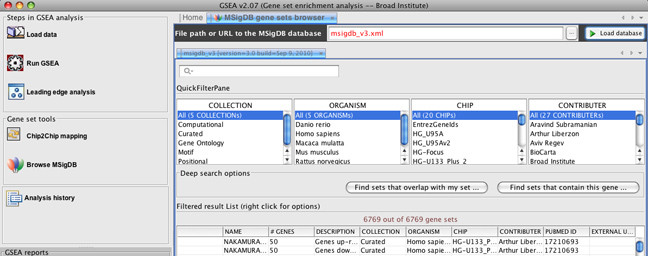
However, if you would like to load v2.5 of MSigDB, you can enter "msigdb_v2.5.xml" in the File path or URL to the MSigDB database and clicking the Load database button.
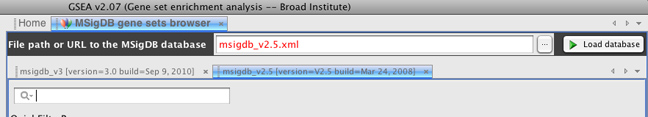
Note in the figure that both the 3.0 and 2.5 versions of MSigDB have been loaded into the browser. You can toggle between these versions by clicking their respective tabs.