Difference between revisions of "Gene Set Pages"
| Line 1: | Line 1: | ||
<p class="MsoListContinue"> </p> | <p class="MsoListContinue"> </p> | ||
| − | <p | + | <p class="MsoNormal" style="">Each gene set in the MSigDB (Molecular Signature Database) is fully described by a gene set card. To review the gene set cards:<br /></p> |
<ol> | <ol> | ||
<li>Click GeneSetCards at the top of this page. The web page displays gene set categories.<br /></li> | <li>Click GeneSetCards at the top of this page. The web page displays gene set categories.<br /></li> | ||
| Line 7: | Line 7: | ||
</ol> | </ol> | ||
Alternatively, from within the GSEA application, use the Browse MSigDB page ([[msigdb_browser]]) to browse gene sets and display gene set cards.<br /><br />[[image:gene_set_card.gif]] | Alternatively, from within the GSEA application, use the Browse MSigDB page ([[msigdb_browser]]) to browse gene sets and display gene set cards.<br /><br />[[image:gene_set_card.gif]] | ||
| − | <p | + | <p class="MsoNormal" style="">Each gene set card contains the following information:</p> |
| − | <p | + | <p class="MsoNormal" style=""><strong style="">Standard name</strong>: Gene set name.</p> |
| − | <p | + | <p class="MsoNormal" style=""><strong style="">LSID</strong>: Internal identifier for the gene set.</p> |
<!--[if !supportLists]--><!--[endif]--> <span style=""></span> | <!--[if !supportLists]--><!--[endif]--> <span style=""></span> | ||
<ul> </ul> | <ul> </ul> | ||
<!--[if !supportLists]--> | <!--[if !supportLists]--> | ||
<ul> </ul> | <ul> </ul> | ||
| − | <p | + | <p class="MsoNormal" style=""><strong style="">Brief description</strong>: A brief description of the gene. This is the description that appears on the Browse MSigDB page of the GSEA application. </p> |
| − | <p | + | <p class="MsoNormal" style=""><strong style="">Category</strong>: The category of the gene set: Positional (C1), Curated (C2), Motif (C3), or Computed (C4). </p> |
<p class="MsoNormal"><strong style="">Sub-category</strong>: The subcategory of the gene set: <br /></p> | <p class="MsoNormal"><strong style="">Sub-category</strong>: The subcategory of the gene set: <br /></p> | ||
<ul> | <ul> | ||
| Line 23: | Line 23: | ||
<li>for C4, always Neighborhood.</li> | <li>for C4, always Neighborhood.</li> | ||
</ul> | </ul> | ||
| − | <p class="MsoNormal"><span style="font-weight: bold;"><strong style="">Full description or Abstract</strong>: A more complete description of the gene set. For example, for gene sets linked to a PubMed entry, this field shows the abstract of that entry. | + | <p class="MsoNormal"><span style="font-weight: bold;"><strong style="">Full description or Abstract</strong>:</span> A more complete description of the gene set. For example, for gene sets linked to a PubMed entry, this field shows the abstract of that entry.</p> |
<p class="MsoNormal"><strong style="">Publication URL</strong>: When available, a link to the source publication. For example, for gene sets linked to a PubMed entry, this is a link to the Entrez record of the source publication. </p> | <p class="MsoNormal"><strong style="">Publication URL</strong>: When available, a link to the source publication. For example, for gene sets linked to a PubMed entry, this is a link to the Entrez record of the source publication. </p> | ||
<p class="MsoNormal"><strong style="">External links</strong>: When available, links to external websites that contain more information about the gene set.</p> | <p class="MsoNormal"><strong style="">External links</strong>: When available, links to external websites that contain more information about the gene set.</p> | ||
| Line 35: | Line 35: | ||
<li><!--[if !supportLists]--><span style=""><span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"></span></span>grp: lists the genes in the gene set in plain text format</li> | <li><!--[if !supportLists]--><span style=""><span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"></span></span>grp: lists the genes in the gene set in plain text format</li> | ||
<li><!--[if !supportLists]--><span style=""><span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"></span></span><!--[endif]-->xml: gene set definition formatted with structure annotation </li> | <li><!--[if !supportLists]--><span style=""><span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"></span></span><!--[endif]-->xml: gene set definition formatted with structure annotation </li> | ||
| − | <li><!--[if !supportLists]--><span style="" | + | <li><!--[if !supportLists]--><span style="">map: lists the genes in the gene set with details of the mapping between the originally reported accessions and their HUGO gene symbols</span></li> |
</ul> | </ul> | ||
<p class="MsoNormal"><strong style="">Genes</strong>: Displays the number of genes (HUGO gene symbols) and accessions (native accessions) in this gene set; if each native accession mapped to a unique HUGO gene symbol, these values are identical. Click the Toggle View link to show/hide the genes in the gene set along with their descriptions and links to external websites for gene-centric information. </p> | <p class="MsoNormal"><strong style="">Genes</strong>: Displays the number of genes (HUGO gene symbols) and accessions (native accessions) in this gene set; if each native accession mapped to a unique HUGO gene symbol, these values are identical. Click the Toggle View link to show/hide the genes in the gene set along with their descriptions and links to external websites for gene-centric information. </p> | ||
Revision as of 10:33, 31 March 2006
Each gene set in the MSigDB (Molecular Signature Database) is fully described by a gene set card. To review the gene set cards:
- Click GeneSetCards at the top of this page. The web page displays gene set categories.
- Click the double arrow next to a category to display the gene sets in that category.
- Click a gene set name to display its gene set card.
Alternatively, from within the GSEA application, use the Browse MSigDB page (msigdb_browser) to browse gene sets and display gene set cards.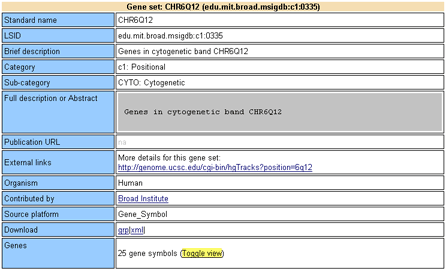
Each gene set card contains the following information:
Standard name: Gene set name.
LSID: Internal identifier for the gene set.
Brief description: A brief description of the gene. This is the description that appears on the Browse MSigDB page of the GSEA application.
Category: The category of the gene set: Positional (C1), Curated (C2), Motif (C3), or Computed (C4).
Sub-category: The subcategory of the gene set:
- for C1, always Cytogenic;
- for C2, Animal Model, Clinical, Experimental pertuberation, Expert curation, Misc, Ontology, Pathway database, or Review paper;
- for C3, Representative motifs or Transfac; and
- for C4, always Neighborhood.
Full description or Abstract: A more complete description of the gene set. For example, for gene sets linked to a PubMed entry, this field shows the abstract of that entry.
Publication URL: When available, a link to the source publication. For example, for gene sets linked to a PubMed entry, this is a link to the Entrez record of the source publication.
External links: When available, links to external websites that contain more information about the gene set.
Keywords & MeSH headings: Keywords derived from MeSH National Library of Medicine and/or manually entered.
Organism: The organism that the experiment was conducted in (generic for canonical pathways and ontologies).
Contributed by: Name of the person (or external database; for example, KEGG) who collected the gene set.
Source platform: Microarray platform that was used for expression profiling (for example, Affymetrix HU6800). Gene_Symbol is used if the gene set was not tied to a specific experiment (for example, for ontologies or a gene set from a review article).
Download: Links to download the gene set in different formats:
- grp: lists the genes in the gene set in plain text format
- xml: gene set definition formatted with structure annotation
- map: lists the genes in the gene set with details of the mapping between the originally reported accessions and their HUGO gene symbols
Genes: Displays the number of genes (HUGO gene symbols) and accessions (native accessions) in this gene set; if each native accession mapped to a unique HUGO gene symbol, these values are identical. Click the Toggle View link to show/hide the genes in the gene set along with their descriptions and links to external websites for gene-centric information.