Difference between revisions of "Gene Set Pages"
| Line 1: | Line 1: | ||
<p class="MsoListContinue"> </p> | <p class="MsoListContinue"> </p> | ||
| − | <p | + | <p class="MsoNormal" style="">Each gene set in the MSigDB (Molecular Signature Database) is fully described by a gene set card. To review the gene set cards:<br /> |
| + | </p> | ||
<ol> | <ol> | ||
| − | <li>Click GeneSetCards at the top of this page. The web page displays gene set categories.<br /></li> | + | <li>Click GeneSetCards at the top of this page. The web page displays gene set categories.<br /> |
| + | </li> | ||
<li>Click the double arrow next to a category to display the gene sets in that category.</li> | <li>Click the double arrow next to a category to display the gene sets in that category.</li> | ||
<li>Click a gene set name to display its gene set card.</li> | <li>Click a gene set name to display its gene set card.</li> | ||
</ol> | </ol> | ||
| − | Alternatively, from within the GSEA application, use the Browse MSigDB page to browse gene sets and display gene set cards. For more information, see the [http://www.broad.mit.edu/cancer/software/ | + | Alternatively, from within the GSEA application, use the Browse MSigDB page to browse gene sets and display gene set cards. For more information, see the [http://www.broad.mit.edu/cancer/software/gsea/doc/GSEAUserGuideFrame.html?Browse_MSigDB_Page Browse MSigDB Page] of the <span style="font-style: italic;">GSEA User Guide</span>.<br /> |
| − | <p | + | <br /> |
| − | <p | + | [[image:gene_set_card.gif]] |
| − | <p | + | <p class="MsoNormal" style="">Each gene set card contains the following information:</p> |
| − | <!--[if !supportLists]--><!--[endif]--> | + | <p class="MsoNormal" style=""><strong style="">Standard name</strong>: Gene set name.</p> |
| − | + | <p class="MsoNormal" style=""><strong style="">LSID</strong>: Internal identifier for the gene set.</p> | |
| − | + | <!--[if !supportLists]--><!--[endif]--> <!--[if !supportLists]--> | |
| − | + | <p class="MsoNormal" style=""><strong style="">Brief description</strong>: A brief description of the gene. This is the description that appears on the Browse MSigDB page of the GSEA application. </p> | |
| − | + | <p class="MsoNormal" style=""><strong style="">Category</strong>: The category of the gene set: Positional (C1), Curated (C2), Motif (C3), or Computed (C4). </p> | |
| − | + | <p class="MsoNormal"><strong style="">Sub-category</strong>: The subcategory of the gene set: <br /> | |
| − | + | </p> | |
| − | + | <ul> | |
| − | + | <li>for C1, always Cytogenic; <br /> | |
| − | + | </li> | |
| − | + | <li>for C2, Animal Model, Clinical, Experimental pertuberation, Expert curation, Misc, Ontology, Pathway database, or Review paper; <br /> | |
| − | + | </li> | |
| − | + | <li>for C3, Representative motifs or Transfac; and <br /> | |
| − | + | </li> | |
| − | + | <li>for C4, always Neighborhood.</li> | |
| − | + | </ul> | |
| − | + | <p class="MsoNormal"><span style="font-weight: bold;"><strong style="">Full description or Abstract</strong>:</span> A more complete description of the gene set. For example, for gene sets linked to a PubMed entry, this field shows the abstract of that entry.</p> | |
| − | + | <p class="MsoNormal"><strong style="">Publication URL</strong>: When available, a link to the source publication. For example, for gene sets linked to a PubMed entry, this is a link to the Entrez record of the source publication. </p> | |
| − | + | <p class="MsoNormal"><strong style="">External links</strong>: When available, links to external websites that contain more information about the gene set.</p> | |
| − | + | <p class="MsoNormal"><strong style="">Keywords & MeSH headings</strong>: Keywords derived from MeSH [http://www.nlm.nih.gov/mesh/MBrowser.html National Library of Medicine] and/or manually entered.</p> | |
| − | + | <p class="MsoNormal"><strong style="">Organism</strong>: The organism that the experiment was conducted in (<em>generic</em> for canonical pathways and ontologies).</p> | |
| − | + | <p class="MsoNormal"><strong style="">Contributed by</strong>: Name of the person (or external database; for example, KEGG) who collected the gene set.</p> | |
| − | + | <p class="MsoNormal"><strong style="">Source platform</strong>: Microarray platform that was used for expression profiling (for example, Affymetrix HU6800). Gene_Symbol is used if the gene set was not tied to a specific experiment (for example, for ontologies or a gene set from a review article).</p> | |
| − | + | <p class="MsoNormal"><strong style="">Download</strong>: Links to download the gene set in different formats:<br /> | |
| − | + | </p> | |
| − | + | <p class="MsoNormal"> </p> | |
| − | + | <ul> | |
| − | + | <li><!--[if !supportLists]--><span style="" />grp: lists the genes in the gene set in plain text format</li> | |
| + | <li><!--[if !supportLists]--><span style="" /><!--[endif]-->xml: gene set definition formatted with structure annotation </li> | ||
| + | <li><!--[if !supportLists]--><span style="">map: lists the genes in the gene set with details of the mapping between the originally reported accessions and their HUGO gene symbols</span></li> | ||
| + | </ul> | ||
| + | <p class="MsoNormal"><strong style="">Genes</strong>: Displays the number of genes (HUGO gene symbols) and accessions (native accessions) in this gene set; if each native accession mapped to a unique HUGO gene symbol, these values are identical. Click the Toggle View link to show/hide the genes in the gene set along with their descriptions and links to external websites for gene-centric information. </p> | ||
Revision as of 14:42, 10 January 2007
Each gene set in the MSigDB (Molecular Signature Database) is fully described by a gene set card. To review the gene set cards:
- Click GeneSetCards at the top of this page. The web page displays gene set categories.
- Click the double arrow next to a category to display the gene sets in that category.
- Click a gene set name to display its gene set card.
Alternatively, from within the GSEA application, use the Browse MSigDB page to browse gene sets and display gene set cards. For more information, see the Browse MSigDB Page of the GSEA User Guide.
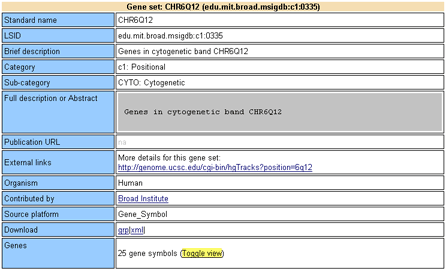
Each gene set card contains the following information:
Standard name: Gene set name.
LSID: Internal identifier for the gene set.
Brief description: A brief description of the gene. This is the description that appears on the Browse MSigDB page of the GSEA application.
Category: The category of the gene set: Positional (C1), Curated (C2), Motif (C3), or Computed (C4).
Sub-category: The subcategory of the gene set:
- for C1, always Cytogenic;
- for C2, Animal Model, Clinical, Experimental pertuberation, Expert curation, Misc, Ontology, Pathway database, or Review paper;
- for C3, Representative motifs or Transfac; and
- for C4, always Neighborhood.
Full description or Abstract: A more complete description of the gene set. For example, for gene sets linked to a PubMed entry, this field shows the abstract of that entry.
Publication URL: When available, a link to the source publication. For example, for gene sets linked to a PubMed entry, this is a link to the Entrez record of the source publication.
External links: When available, links to external websites that contain more information about the gene set.
Keywords & MeSH headings: Keywords derived from MeSH National Library of Medicine and/or manually entered.
Organism: The organism that the experiment was conducted in (generic for canonical pathways and ontologies).
Contributed by: Name of the person (or external database; for example, KEGG) who collected the gene set.
Source platform: Microarray platform that was used for expression profiling (for example, Affymetrix HU6800). Gene_Symbol is used if the gene set was not tied to a specific experiment (for example, for ontologies or a gene set from a review article).
Download: Links to download the gene set in different formats:
- grp: lists the genes in the gene set in plain text format
- xml: gene set definition formatted with structure annotation
- map: lists the genes in the gene set with details of the mapping between the originally reported accessions and their HUGO gene symbols
Genes: Displays the number of genes (HUGO gene symbols) and accessions (native accessions) in this gene set; if each native accession mapped to a unique HUGO gene symbol, these values are identical. Click the Toggle View link to show/hide the genes in the gene set along with their descriptions and links to external websites for gene-centric information.