Difference between revisions of "New Page To Test DB Fix"
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | Each gene set in the MSigDB (Molecular Signature Database) is fully described by a gene set card. From this web site, use the [http://gsea.broad.mit.edu:8080/cancer/msigdb/ MSigDB page] to find a gene set. Click the gene set name to display its gene set card. Alternatively, from within the GSEA application, use the [http://www.broad.mit.edu/cancer/software/gsea/doc/GSEAUserGuideFrame.html?Browse_MSigDB_Page Browse MSigDB page] to browse gene sets and display gene set cards. <br /> | |
| + | <br /> | ||
| + | [[image:gene_set_card.gif]] **update** this is an edit<br /> | ||
| + | <br /> | ||
| + | [[image:msigdb_local.gif]]<br /> | ||
| + | <br /> | ||
| + | Each gene set card contains the following information:<br /> | ||
| + | <br /> | ||
| + | <strong> Brief description</strong>: A brief description of the gene. This is the description that appears on the Browse MSigDB page of the GSEA application.<br /> | ||
| + | <br /> | ||
| + | <strong> Full description or abstract</strong>: A more complete description of the gene set. For example, for gene sets linked to a PubMed entry, this field shows the abstract of that entry.<br /> | ||
| + | <br /> | ||
| + | <strong>Collection</strong>: The collection that contains the gene set: Positional (C1), Curated (C2), Motif (C3), or Computed (C4). See the [http://gsea.broad.mit.edu:8080/cancer/msigdb/aboutCollections.jsp MSigDB Collections page] for a description of each collection.<br /> | ||
| + | <br /> | ||
| + | <strong>Source publication</strong>: When available, a link to the source publication followed by a list of its authors. <br /> | ||
| + | <br /> | ||
| + | <strong> Related gene sets</strong>: Other gene sets (if any) from this publication. Other gene sets (if any) from publications by one or more of these authors.<br /> | ||
| + | <br /> | ||
| + | <strong>External links</strong>: When available, links to external websites that contain more information about the gene set.<br /> | ||
| + | <br /> | ||
| + | <strong>Organism</strong>: The organism that the experiment was conducted in (generic for canonical pathways and ontologies).<br /> | ||
| + | <br /> | ||
| + | <strong> Contributed by</strong>: Name of the person (or external database; for example, KEGG) who collected the gene set.<br /> | ||
| + | <br /> | ||
| + | <strong> Source platform</strong>: Microarray platform that was used for expression profiling (for example, Affymetrix HU6800). Gene_Symbol is used if the gene set was not tied to a specific experiment (for example, for ontologies or a gene set from a review article).<br /> | ||
| + | <br /> | ||
| + | <strong> Download gene set</strong>: Links to download the gene set in different formats: grp (group), text, gmt, or gmx. For descriptions of each format, see [[Data_formats|Data Formats]].<br /> | ||
| + | <br /> | ||
| + | <strong>Annotate by computing overlaps</strong>: Displays the overlap (if any) between this gene set and the gene sets in the selected collection.<br /> | ||
| + | <br /> | ||
| + | <strong>Compendia expression profiles</strong>: Displays a heat map showing how the genes in this gene set are expressed in the selected expression dataset.<br /> | ||
| + | <br /> | ||
| + | <strong>Advanced query</strong>: Copies the genes in this gene set to the annotations page, where you can modify and/or further explore the gene set.<br /> | ||
<br /> | <br /> | ||
| − | + | <strong>Gene families</strong>: Displays a functional overview of this gene set by categorizing its members into a small number of gene families, where each gene family describes a collection of proteins that share a common feature such as homology or biochemical activity.<br /> | |
| − | |||
| − | |||
| − | |||
| − | |||
<br /> | <br /> | ||
| − | + | <strong>Show x genes</strong>: Specifies the number (<strong>x</strong>) of genes in this gene set. Click the Show (Hide) link show (hide) to list each gene (HUGO gene symbols) followed by links to external websites for gene-centric information. <br /> | |
<br /> | <br /> | ||
| − | < | + | <hr width="100%" size="2" /> |
| − | |||
| − | |||
<br /> | <br /> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<br /> | <br /> | ||
| − | + | Old Documentation Home page<br /> | |
| − | |||
| − | |||
<br /> | <br /> | ||
| − | + | <a href="http://www.broad.mit.edu/gsea/">GSEA Home</a> | <a href="../../software/software_index.html">Software</a> | <a href="../../msigdb/msigdb_index.html">MSigDB</a> | Documentation | <a href="../../resources/resources_index.html">Resources</a> <br /> | |
<br /> | <br /> | ||
| + | <table cellspacing="1" cellpadding="1" border="0" align="" summary="" style="width: 689px; height: 84px;"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td><font size="4">[http://www.broad.mit.edu/cancer/software/gsea/doc/GSEAUserGuideFrame.htm User Guide]</font></td> | ||
| + | <td><font size="4">[[Release_Notes|GSEA Release Notes]]</font></td> | ||
| + | <td><font size="4">[[Gsea_Algorithm|GSEA Algorithm]]</font></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><font size="4">[http://www.broad.mit.edu/cancer/software/gsea/doc/desktop_tutorial.html Tutorial]</font></td> | ||
| + | <td><font size="4"><font size="4">[[Release_Notes|MSigDB Release Notes]]<br /> | ||
| + | </font></font></td> | ||
| + | <td><font size="4">[http://www.broad.mit.edu/cancer/software/gsea/resources/datasets_index.html Example Datasets]</font></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><font size="4">[[FAQ]]</font></td> | ||
| + | <td><font size="4">[[Gene_set_card_guide|Guide to Gene Set Cards]]</font></td> | ||
| + | <td><font size="4">[[Gsea_papers|GSEA Papers]]</font></td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
<br /> | <br /> | ||
| + | <font size="3">To cite GSEA, please reference Subramanian, Tamayo, et al. ([http://www.pnas.org/cgi/content/abstract/102/43/15545 2005, PNAS 102, 15545-15550]).<br /> | ||
| + | </font><br /> | ||
| + | <font size="4"><font size="3">If you have questions or comments, contact us: [mailto:gsea@broad.mit.edu gsea@broad.mit.edu].<br /> | ||
| + | </font></font><br /> | ||
| | ||
Latest revision as of 10:48, 15 December 2008
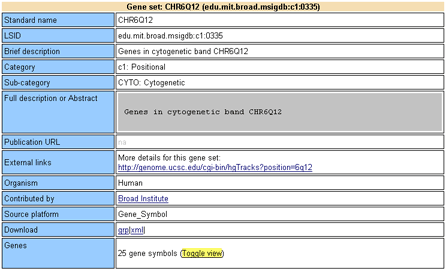
Each gene set in the MSigDB (Molecular Signature Database) is fully described by a gene set card. From this web site, use the MSigDB page to find a gene set. Click the gene set name to display its gene set card. Alternatively, from within the GSEA application, use the Browse MSigDB page to browse gene sets and display gene set cards.
 **update** this is an edit
**update** this is an edit

Each gene set card contains the following information:
Brief description: A brief description of the gene. This is the description that appears on the Browse MSigDB page of the GSEA application.
Full description or abstract: A more complete description of the gene set. For example, for gene sets linked to a PubMed entry, this field shows the abstract of that entry.
Collection: The collection that contains the gene set: Positional (C1), Curated (C2), Motif (C3), or Computed (C4). See the MSigDB Collections page for a description of each collection.
Source publication: When available, a link to the source publication followed by a list of its authors.
Related gene sets: Other gene sets (if any) from this publication. Other gene sets (if any) from publications by one or more of these authors.
External links: When available, links to external websites that contain more information about the gene set.
Organism: The organism that the experiment was conducted in (generic for canonical pathways and ontologies).
Contributed by: Name of the person (or external database; for example, KEGG) who collected the gene set.
Source platform: Microarray platform that was used for expression profiling (for example, Affymetrix HU6800). Gene_Symbol is used if the gene set was not tied to a specific experiment (for example, for ontologies or a gene set from a review article).
Download gene set: Links to download the gene set in different formats: grp (group), text, gmt, or gmx. For descriptions of each format, see Data Formats.
Annotate by computing overlaps: Displays the overlap (if any) between this gene set and the gene sets in the selected collection.
Compendia expression profiles: Displays a heat map showing how the genes in this gene set are expressed in the selected expression dataset.
Advanced query: Copies the genes in this gene set to the annotations page, where you can modify and/or further explore the gene set.
Gene families: Displays a functional overview of this gene set by categorizing its members into a small number of gene families, where each gene family describes a collection of proteins that share a common feature such as homology or biochemical activity.
Show x genes: Specifies the number (x) of genes in this gene set. Click the Show (Hide) link show (hide) to list each gene (HUGO gene symbols) followed by links to external websites for gene-centric information.
Old Documentation Home page
<a href="http://www.broad.mit.edu/gsea/">GSEA Home</a> | <a href="../../software/software_index.html">Software</a> | <a href="../../msigdb/msigdb_index.html">MSigDB</a> | Documentation | <a href="../../resources/resources_index.html">Resources</a>
| User Guide | GSEA Release Notes | GSEA Algorithm |
| Tutorial | MSigDB Release Notes |
Example Datasets |
| FAQ | Guide to Gene Set Cards | GSEA Papers |
To cite GSEA, please reference Subramanian, Tamayo, et al. (2005, PNAS 102, 15545-15550).
If you have questions or comments, contact us: gsea@broad.mit.edu.